Tools
We are developing open source tools for research and education at the intersection of molecular/systems modeling, machine learning and optimization. These tools include foundational resources that enable therapeutic design and application specific models.
Research
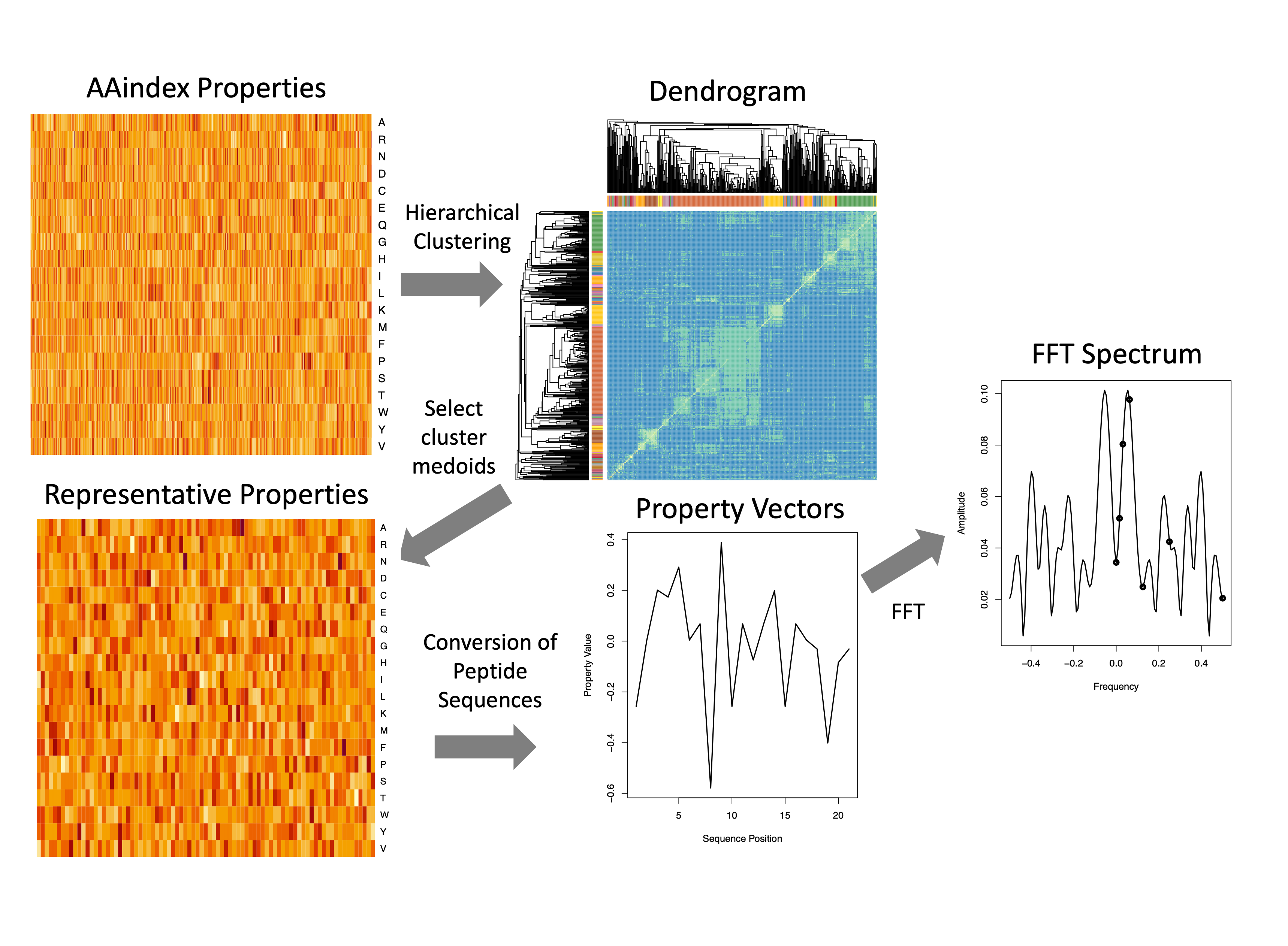
Python package for developing and analyzing machine learned models of protein function and properties that leverage periodicities in physicochemical properties along protein sequences.
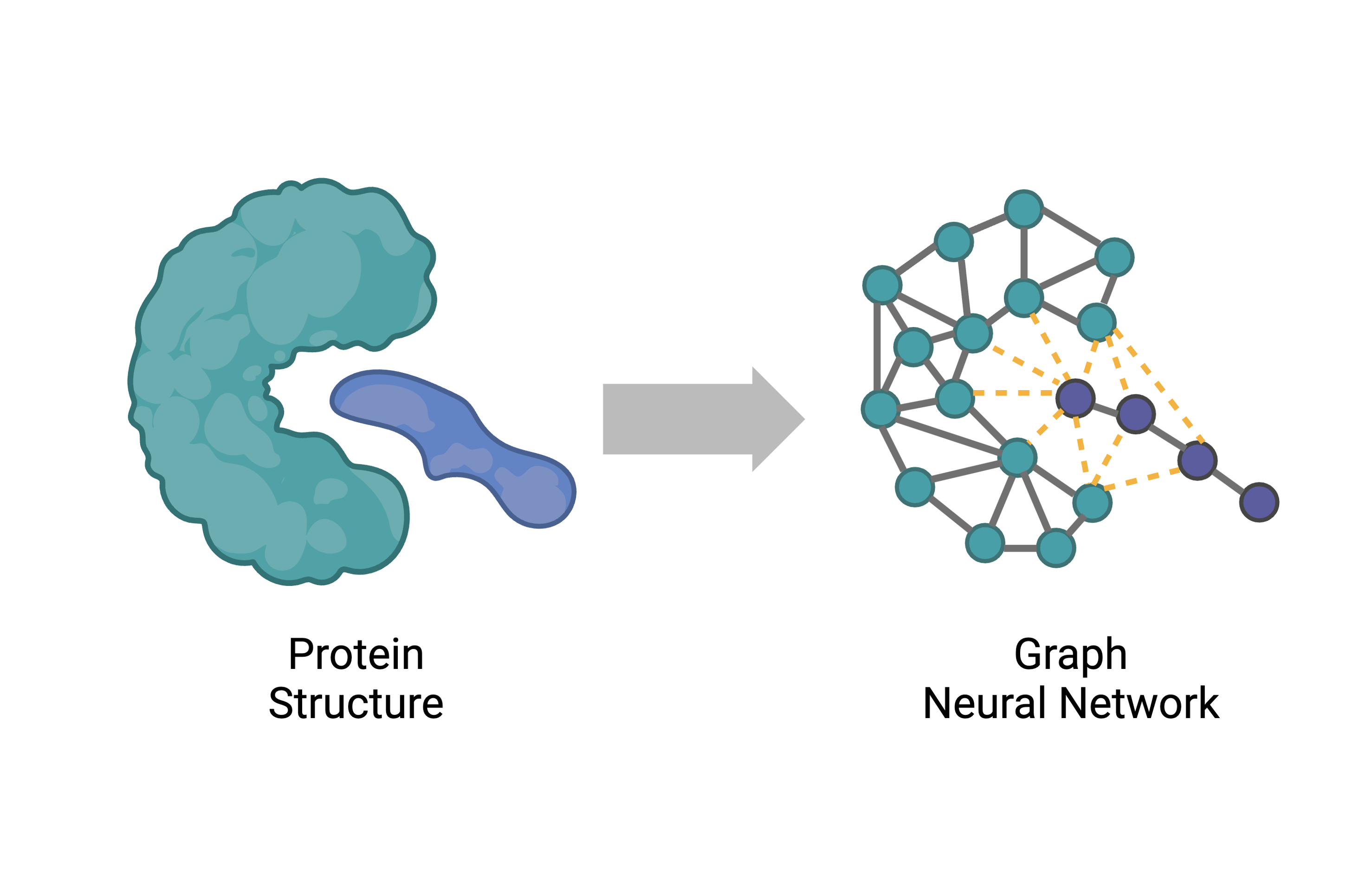
Provides basic tools for manipulating PDB files, including visualization through dash.bio, and provides a framework for developing Graph Neural Networks based portein structural data.
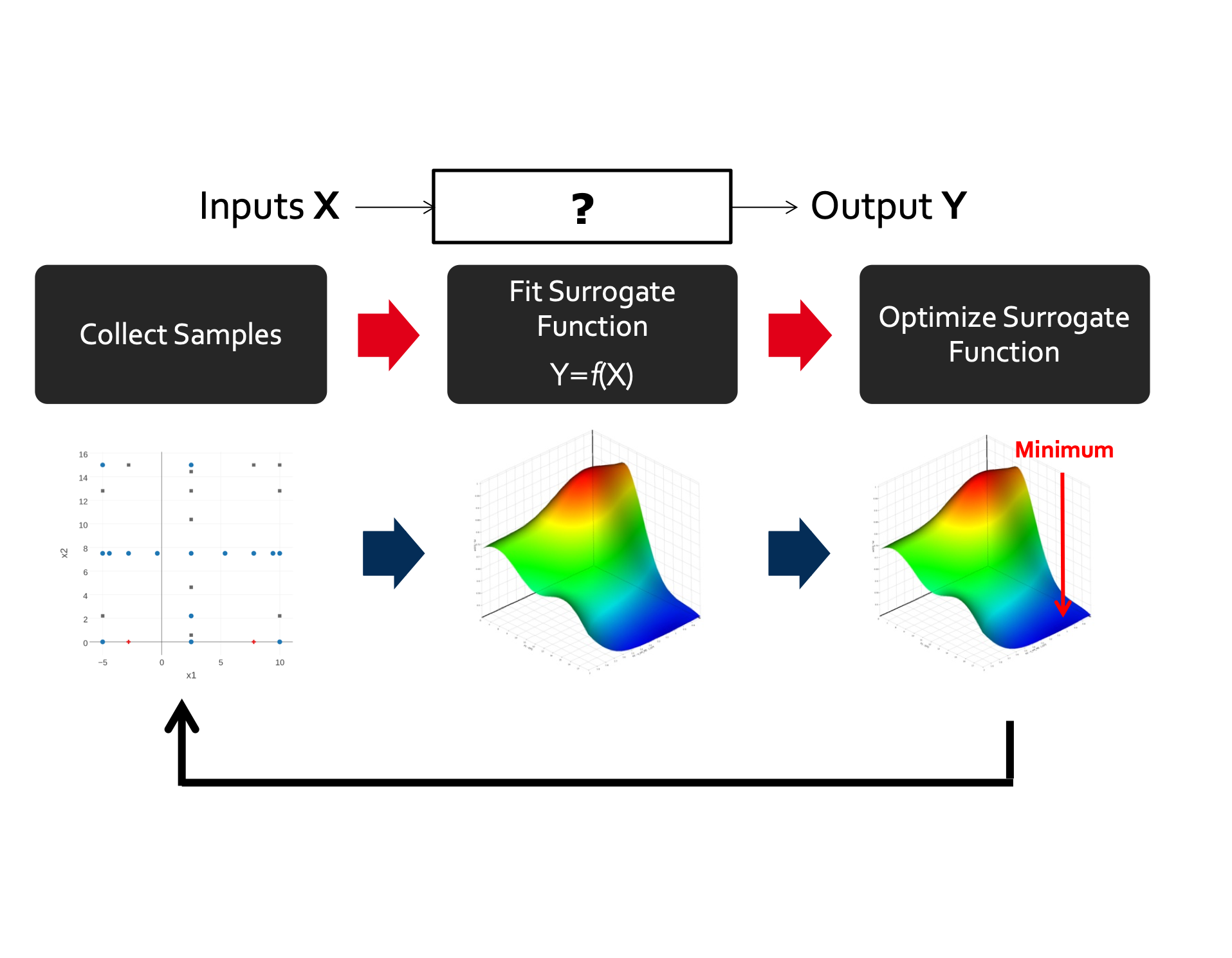
Work in collaboration with Mike Howard (Auburn University) aimed at developing tools for multi-scale modeling based on Smolyak sparse grids.
Education
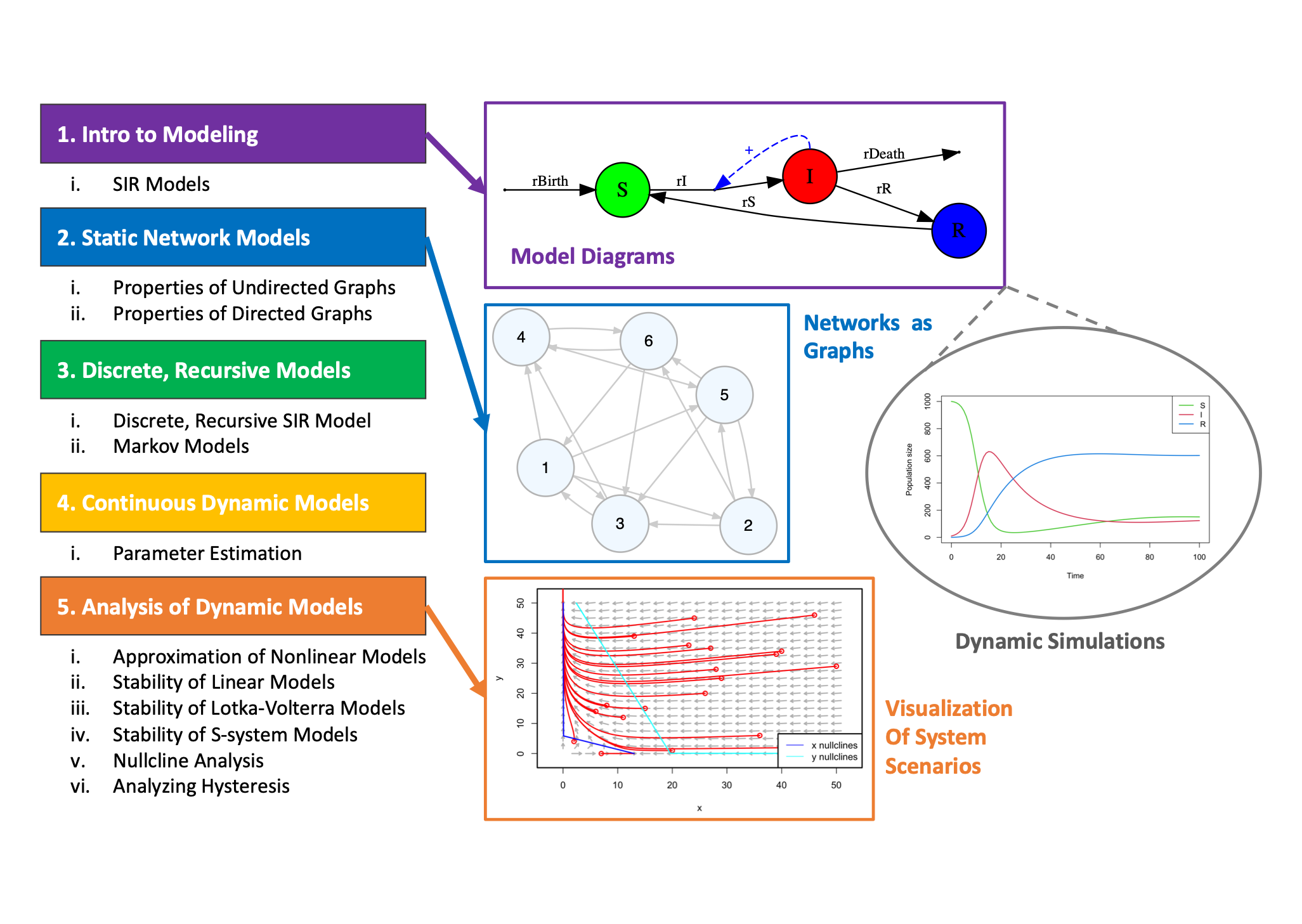
These resources include interactive modules that are generally applicable to learning the basics of systems biology modeling, as well as, coding examples for using R to develop and analyze various types of systems biology models.