Research
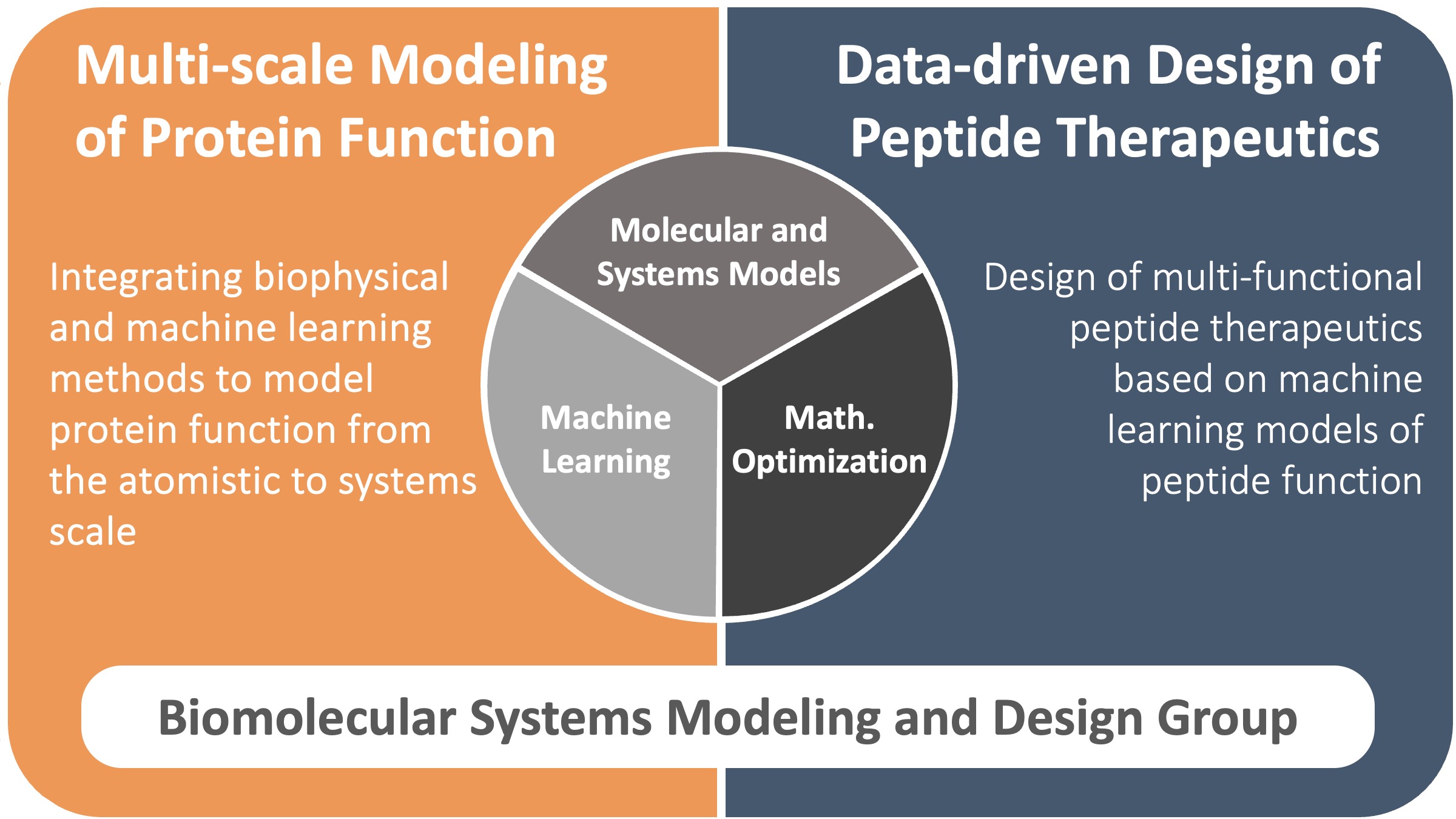
There is serious concern that the continued increase in drug development costs is unsustainable and will lead to further increases in already exorbitant drug prices, highlighting the need for a new paradigm for therapeutic discovery. To meet these challenges, we develop tools for therapeutic design to accelerate discovery and enable personalized medicine. Our work integrates molecular and systems modeling, machine learning, and global optimization to develop multi-scale models of biomolecular systems, and then leverages these models for the design of novel therapeutics.
Current Projects
Our work is currently funded by an R35 MIRA ESI award from the National Institute for General Medical Sciences at National Institutes of Health and start-up and seed funding from Auburn University.
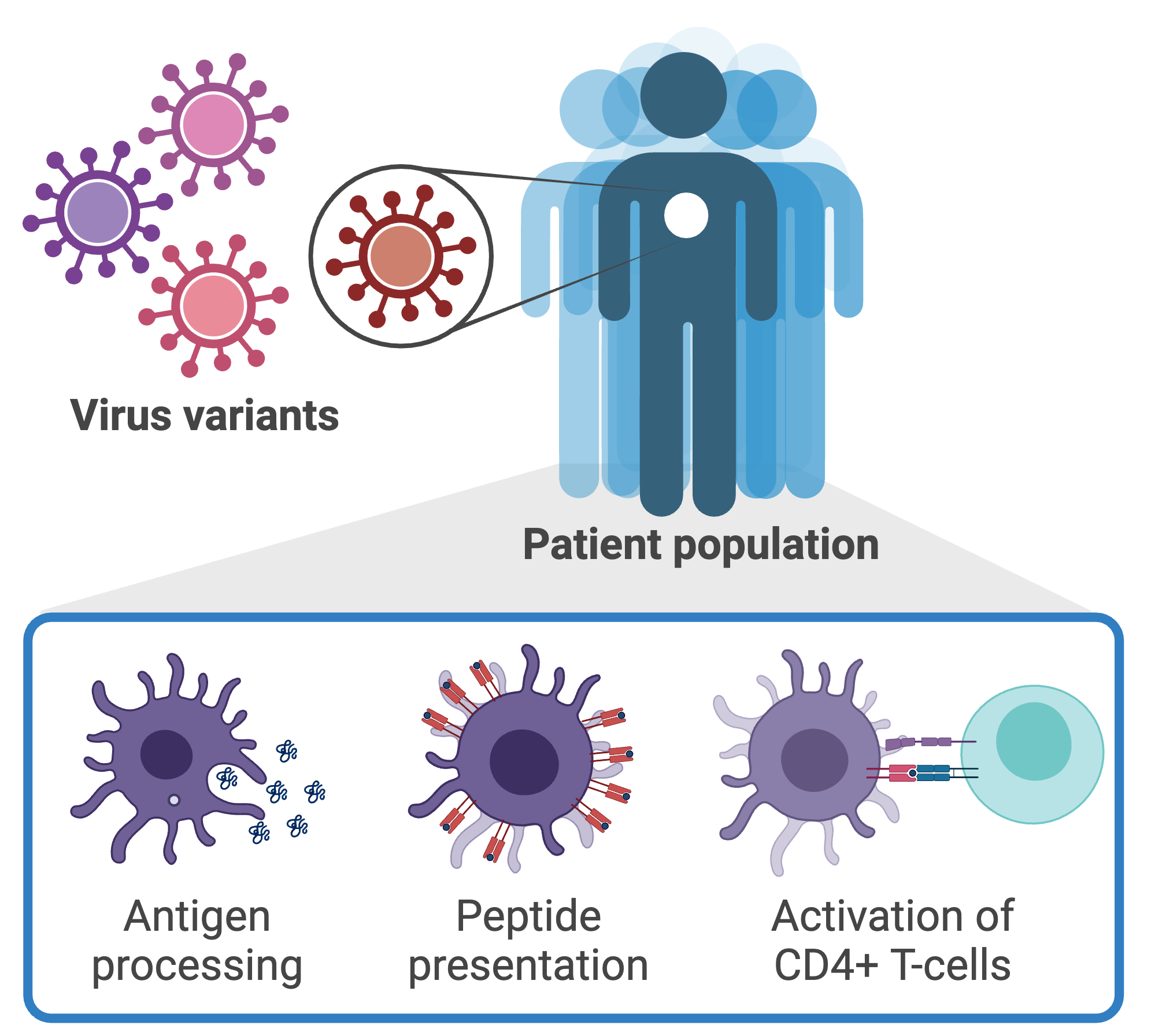
AI-enabled design of personalized vaccines and immunotherapies
Computational methods have emerged as a promising approach for peptide-based vaccine design, where models are used to identify which fragments of the pathogen (epitopes) would serve as a representative element to elicit a robust immune response. Epitope prediction involves predicting the outcomes of antigen processing and presentation given the genetic sequence of a pathogen. The challenges of epitope prediction are compounded further due to the existence of uncertainty caused by genetic variability across pathogen variants and individual to individual. We are developing a computational framework for identifying peptide epitopes that couples biomolecular structure, machine learning, and probabilistic models to account for the effects of genetic mutations on the interactions between the pathogen and host.
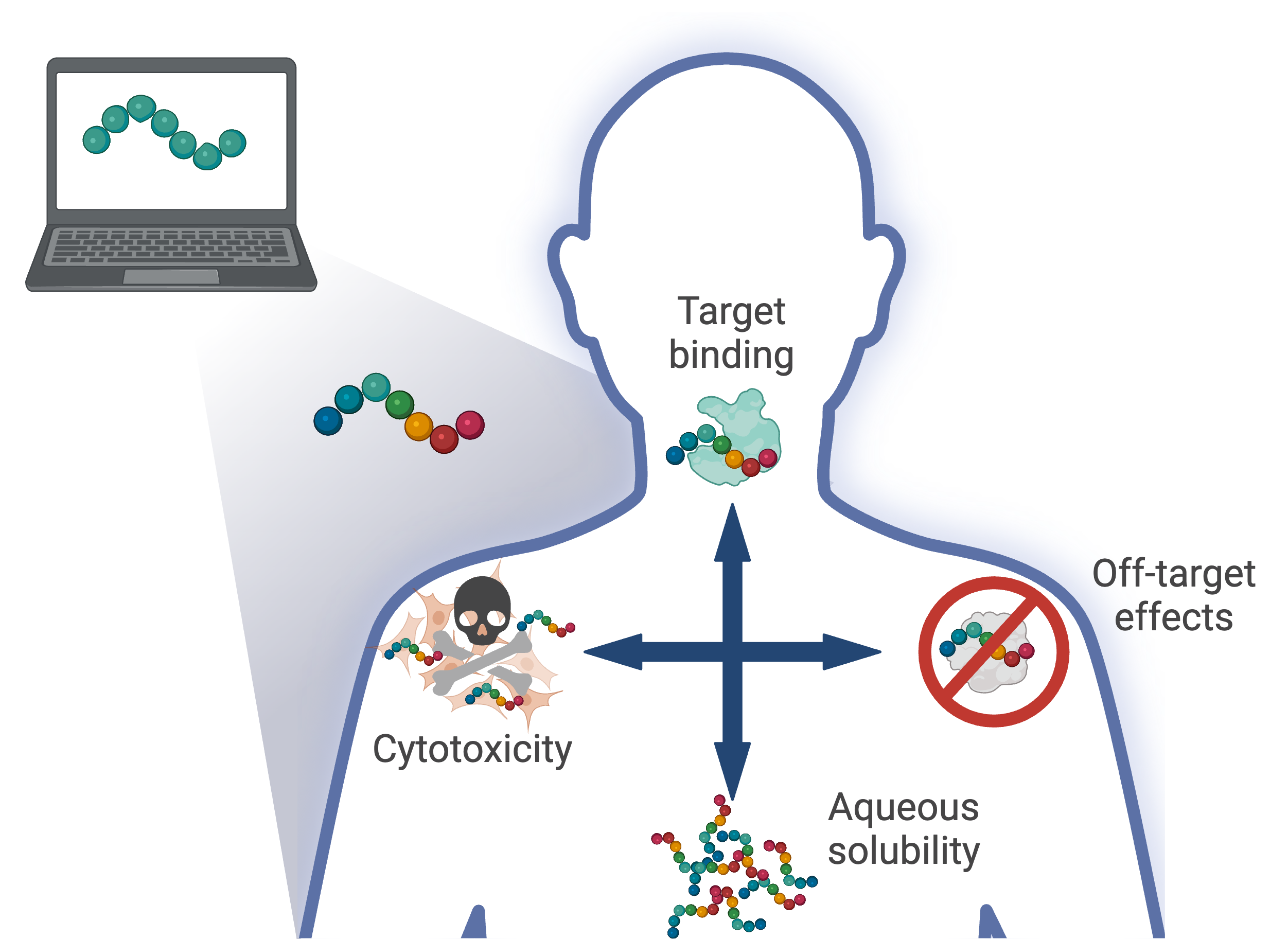
Data-driven design of multi-functional therapeutic peptides
The sequential nature of the drug development pipeline and an inability to predict late-stage failures are often at the heart of long delays and exorbitant costs that have been associated with pharmaceutical development. Integrating a systems perspective with computational molecular design can accelerate the development of novel therapies and reduce late-stage clinical failures through the incorporation of additional design considerations into the design procedure. We are developing a data-driven multi-objective design framework that can be extended to incorporate potentially many design considerations that will be capable of designing peptides that are multi-functional, but specific, while also possessing improved functional properties.
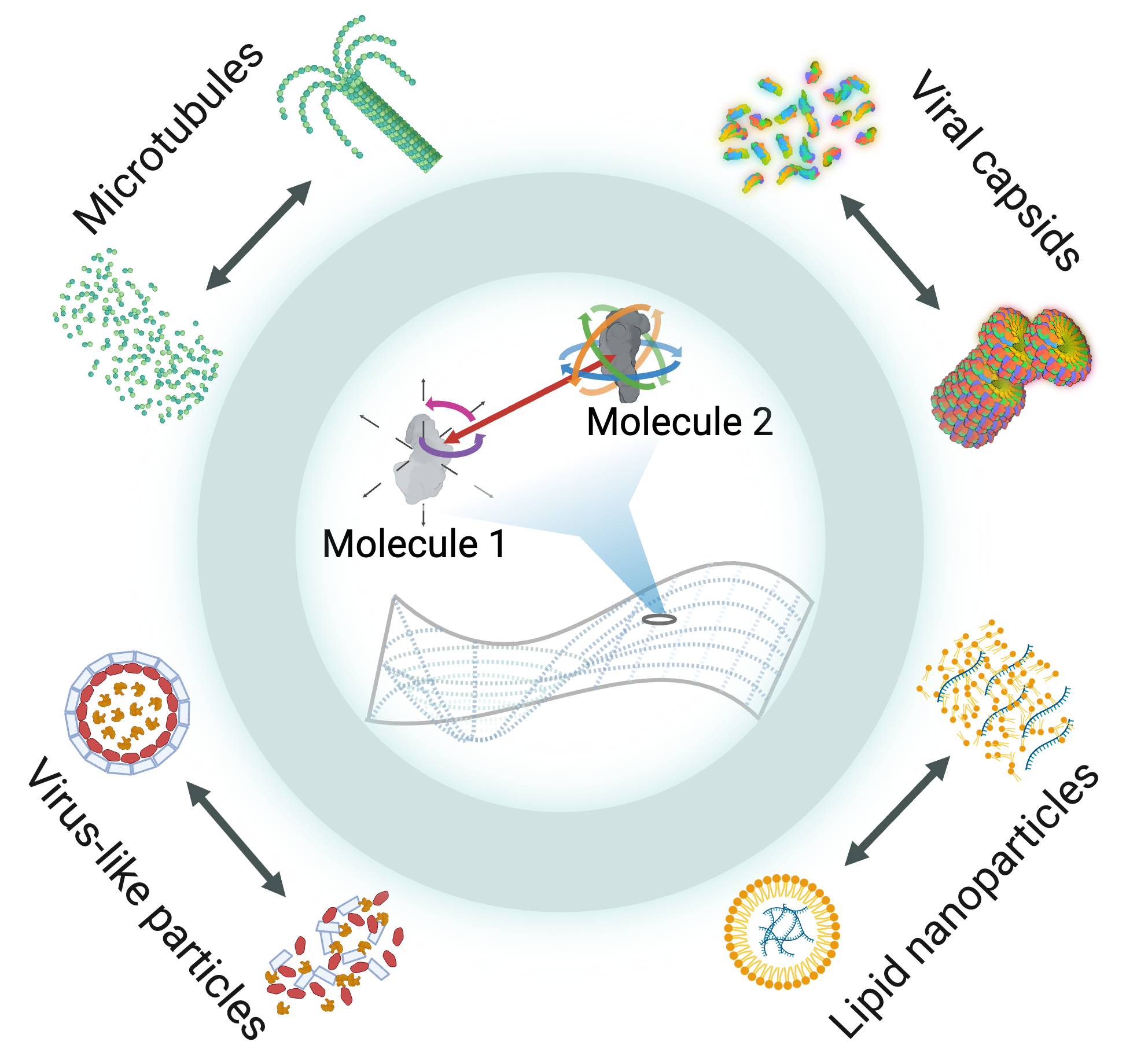
Multi-scale modeling, simulations, and design of
biomolecular self-assembly
Biomolecular self-assembly is fundamental to numerous aspects of biology and developing accurate models could advance many fundamental research areas and technologies in medicine. Potential applications include understanding complex structures such as microtubules, viruses, and biomolecular condensates, as well as designing nanomaterials like lipid nanoparticles or virus-like particles. However, self-assembly is an inherently mesoscale phenomena that occurs at disparate length and time scales, motivating the need for computationally efficient mesoscale models of biomolecules. In collaboration with Michael Howard at Auburn University, we are developing a computational framework for building ultra-coarse models of biomolecular interactions that uses machine learning to approximate mesoscale interactions based on atomistic simulations.